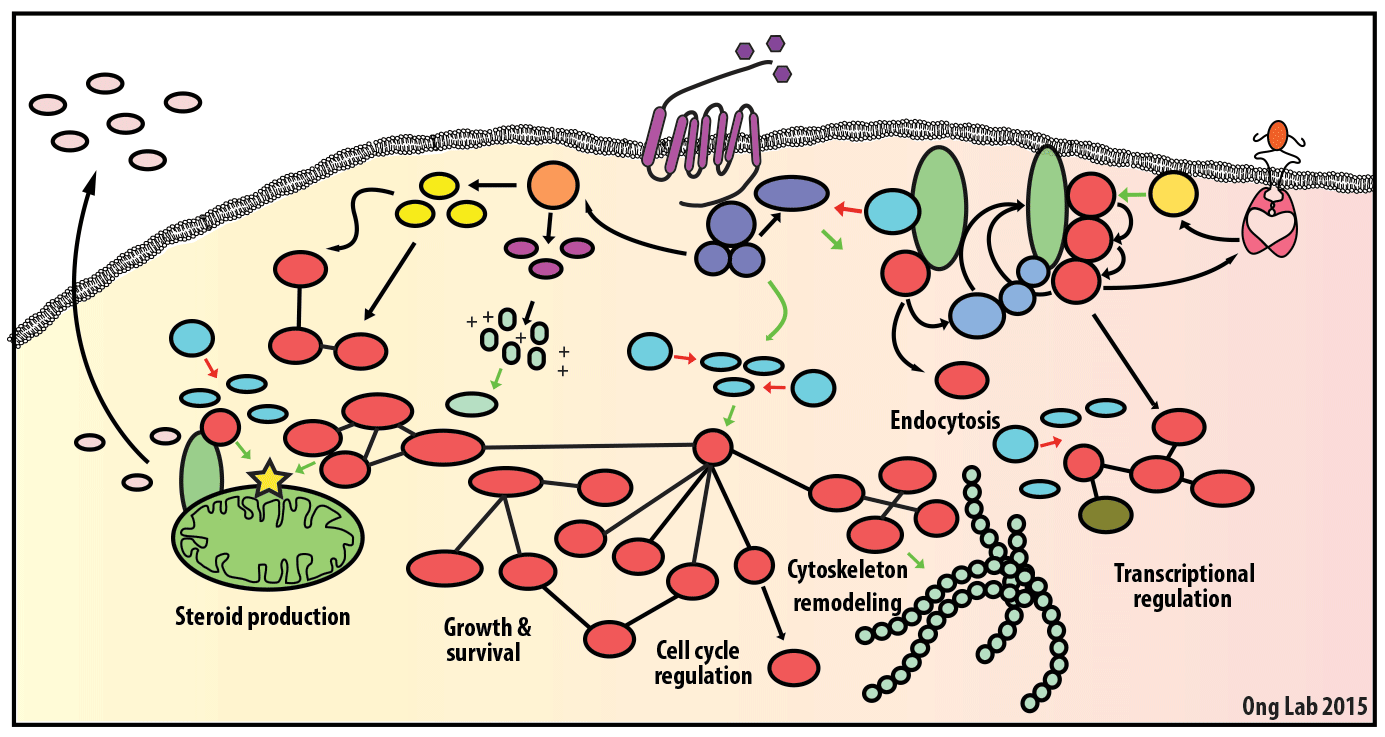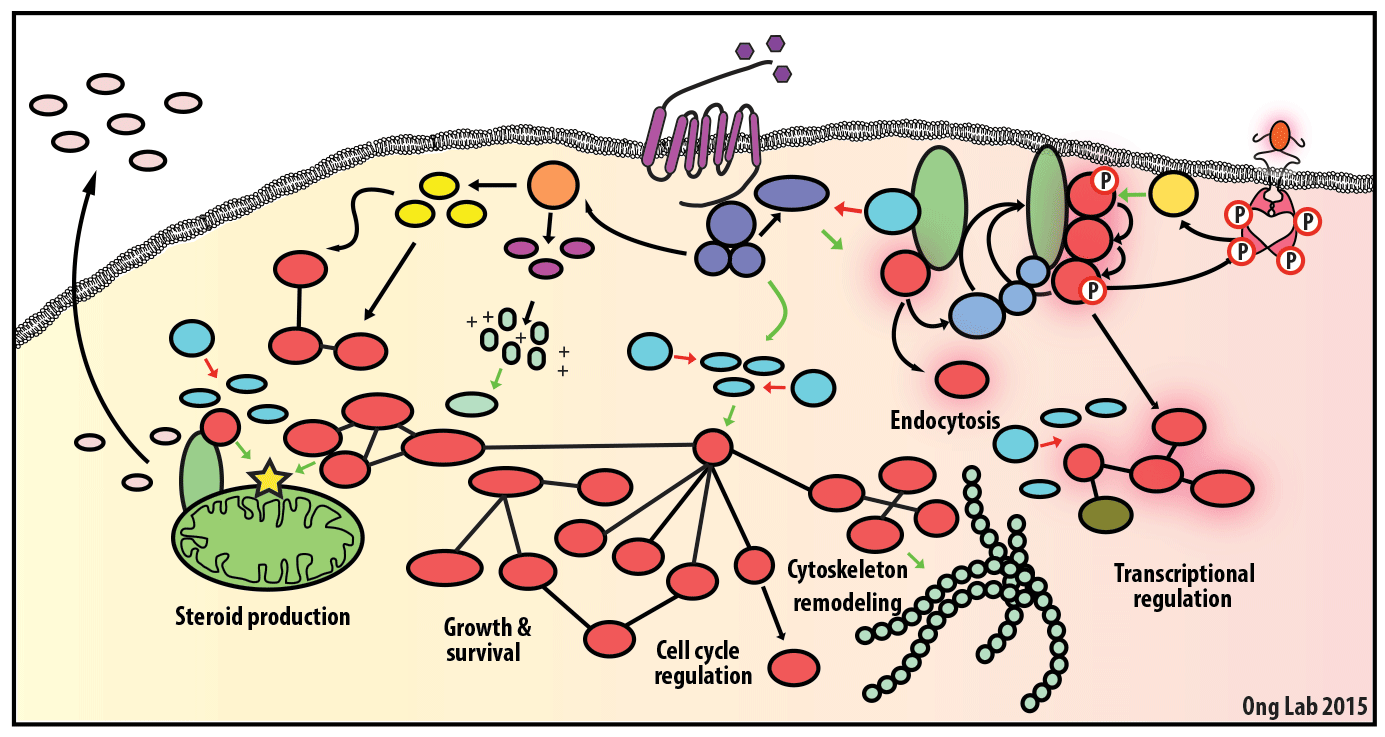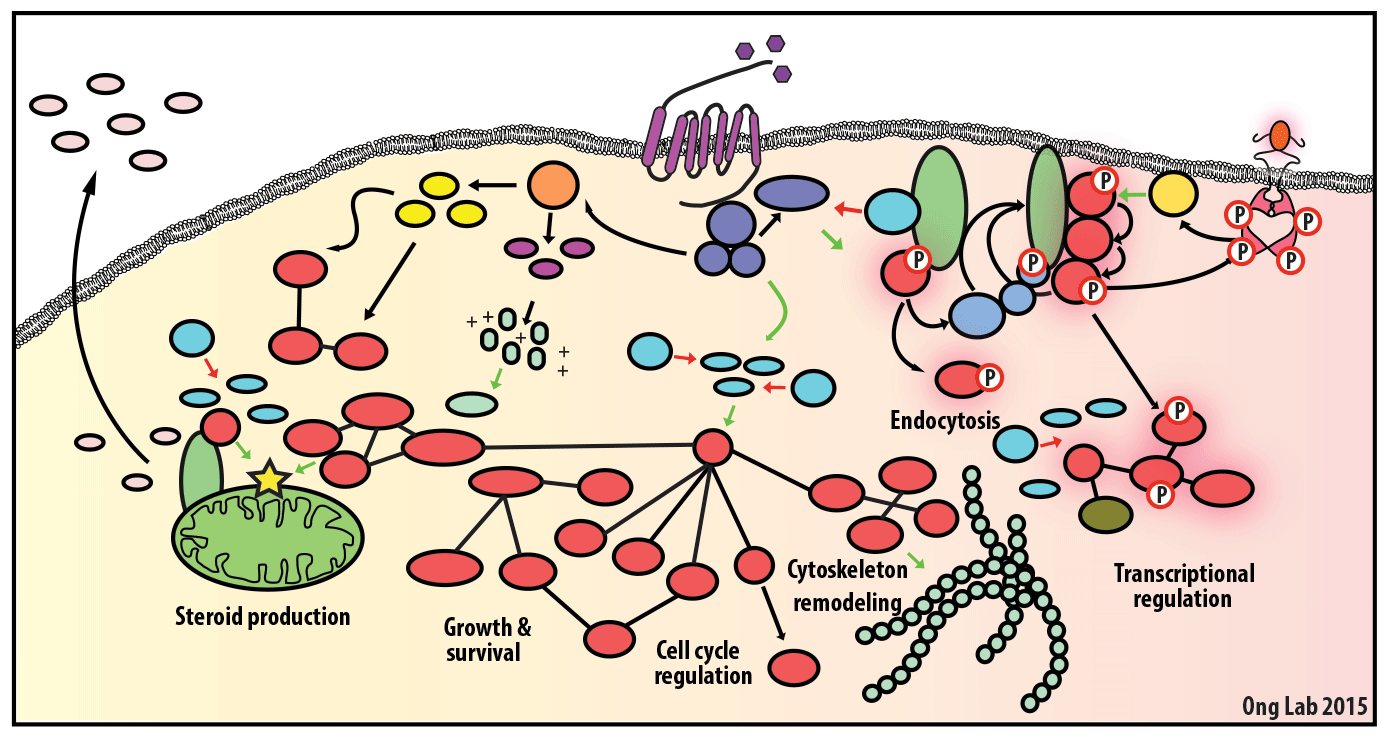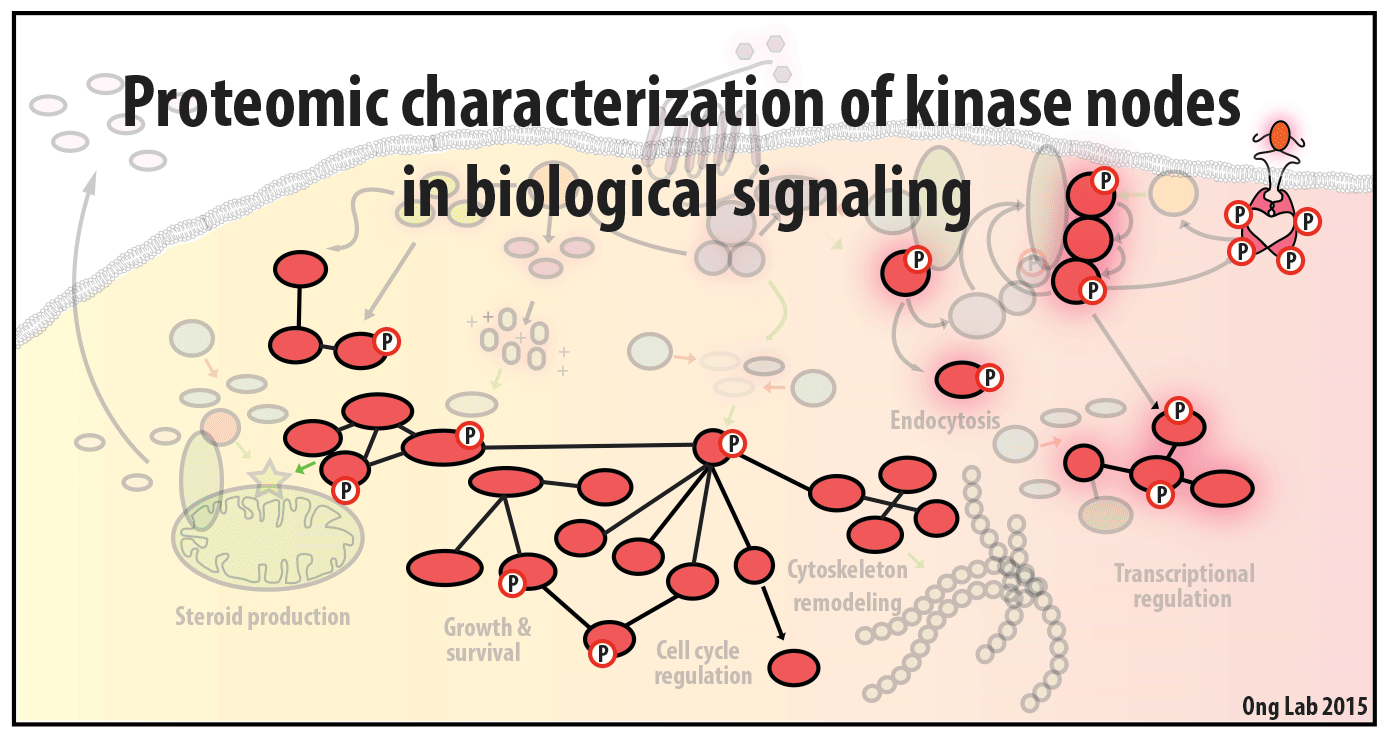
We use mass spectrometry-based analyses to study cell signaling. We have focused on the analyses of kinases, key regulatory nodes in many cellular signaling pathways. To enhance our ability to study these kinases, we use synthesize our own affinity reagents1 that use immobilized non-selective kinase inhibitors to selectively enrich ~70% of the kinases known in the human genome (these affinity reagents are also known as Kinobeads2 or mixed inhibitor beads, MIBs3). This allows us to compare kinase expression and activity between different cellular states, using quantitative proteomics approaches like SILAC.
View our Selected publications for a curated list of work from the lab.
References:
1. Golkowski M, Vidadala RS, Lombard CK, Suh HW, Maly DJ, Ong SE. Kinobead and Single-Shot LC-MS Profiling Identifies Selective PKD Inhibitors. J Proteome Res. 2017;16(3):1216-27. doi: 10.1021/acs.jproteome.6b00817. PubMed PMID: 28102076.
2. Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035-44. doi: 10.1038/nbt1328. PubMed PMID: 17721511.
3. Cooper MJ, Cox NJ, Zimmerman EI, Dewar BJ, Duncan JS, Whittle MC, Nguyen TA, Jones LS, Ghose Roy S, Smalley DM, Kuan PF, Richards KL, Christopherson RI, Jin J, Frye SV, Johnson GL, Baldwin AS, Graves LM. Application of multiplexed kinase inhibitor beads to study kinome adaptations in drug-resistant leukemia. PLoS One. 2013;8(6):e66755. doi: 10.1371/journal.pone.0066755. PubMed PMID: 23826126; PMCID: PMC3691232.